Can you learn to diagnose the severity of aortic stenosis (AS), a common valve disease, from ultrasound images of the heart (echocardiograms)?
The Tufts Medical Echocardiogram Dataset (TMED) is a clinically-motivated benchmark dataset for computer vision and machine learning from limited labeled data. This dataset is designed to be an authentic assessment of semi-supervised learning (SSL) methods that train classifiers from a small, hard-to-acquire labeled dataset and a much larger (but easier to acquire) unlabeled set.
Jump to: Announcements Clinical Motivation Dataset Summary Classification Tasks Bibliography
Announcements
Apr 2023: Methods paper accepted at AISTATS '23
We present a new benchmark for assessing cross-hospital generalization of view-type classifiers that we call Heart2Heart: you can train on TMED then test on data from the UK (Unity) and France (CAMUS). We also have new SSL methods that are effective yet simple: Paper YouTube
Jan 2023: Clinical journal paper accepted at J. Amer. Soc. of Echocardiography
Our JASE '23 paper shows that automatic AS diagnosis is possible for a clinical audience, with external validation Publisher Link PubMed
Nov 18 2022: TMED virtual retreat 2022
See event page for details and how to RSVP to attend!
July 2022: Short paper on TMED-2 will be presented at the DataPerf workshop at ICML 2022
Our short paper is now available as a PDF and 3-minute spotlight video
July 2022: Release of TMED-2 Dataset and Code
Our new expanded dataset, TMED-2, doubles the size of TMED-1, increases resolution to 112x112 pixels, and expands available view labels.
TMED-2 data is available for academic research: Apply for Access LICENSE
TMED-2 code is also available: TMED-2 GitHub repo MIT LICENSE
Apr. 2022: TMED-1 has 60 approved users from 19 countries across 5 continents!
Aug. 2021: Paper accepted at MLHC 2021
Our publication is now available as a PDF and 3-minute spotlight video
July 2021: Release of TMED-1 Dataset and Code
TMED-1 data is available for academic research: Apply for Access LICENSE
TMED-1 code is also available: TMED-1 GitHub repo MIT LICENSE
Clinical motivation
Our motivating task is to improve timely diagnosis and treatment of aortic stenosis (AS), a common degenerative cardiac valve condition. If left untreated, severe AS has lower 5-year survival rates than several metastatic cancers (@howladerSEERCancerStatistics2020; @clarkFiveyearClinicalEconomic2012). With timely diagnosis, AS becomes a treatable condition via surgical or transcatheter aortic valve replacement with very low mortality (@lancellottiOutcomesPatientsAsymptomatic2018).
AS is a particularly important condition where automation holds substantial promise. There is evidence that many patients with severe AS are not treated (@tangContemporaryReasonsClinical2018; @brennanProviderlevelVariabilityTreatment2019) and there are disparities in access to care that must be addressed (@alkhouliRacialDisparitiesUtilization2019). Automated screening for AS can increase referral and treatment rates for patients with this life threatening condition.
We hope this dataset release catalyzes research in two directions:
-
Deployable automatic preliminary screening and early detection of cardiac disease, especially expanding access to patients who live in areas without expert cardiologists (but where ultrasound imaging would still be possible)
-
Improved ML methodology for learning from limited labeled data. For our use case and many others, acquiring appropriate labels from expert clinicians is expensive and time consuming. Our dataset deliberately supports semi-supervised methods that can learn simultaneously from a small labeled dataset and a large unlabeled dataset (much easier to collect).
Dataset Summary
The TMED dataset contains transthoracic echocardiogram (TTE) imagery acquired in the course of routine care consistent with American Society of Echocardiography (ASE) guidelines, all obtained from 2011-2020 at Tufts Medical Center.
When gathering echocardiogram imagery for a patient, a sonographer manipulates a handheld transducer over the patient’s chest, manually choosing different acquisition angles in order to fully assess the heart’s complex anatomy. This imaging process results in multiple cineloop video clips of the heart depicting various anatomical views (see example view types below). We extract one still image from each available video clip, so each patient study is represented in our dataset as multiple images (typically 50-100).
In routine care when images are captured, neither view nor diagnostic labels are immediately captured and stored. View labels are not annotated or stored as part of routine practice. Diagnostic labels for aortic stenosis (AS) - along with many other observations about many aspects of heart health -- are applied hours or days after a study by an expert clinician, who aggregates information from the many videos and images captured by the echocardiogram study. Diagnostic severity ratings are entered some time later into a human readable report document stored within that patient's electronic medical record. Due to logistical reasons it is difficult to easily extract that information into a machine readable format.
We have performed significant annotation effort to gather appropriate view labels for a subset of data, as well as significant manual effort to extract diagnostic labels from existing medical records.
Each view label was provided by experts (board-certified sonographers or cardiologists) specifically for this study using a custom labeling tool. These view types are a subset of the many possible view types, chosen because they are relevant for diagnosing valve diseases like AS.
Each diagnostic label was assigned by a board-certified cardiologist in the course of routine practice when interpreting the echocardiogram to care for the patient. These labels were pulled from the patient's medical record in a manually intensive process.
We have released two versions (TMED-1 and TMED-2). The first represents our original release in July 2021, the second represents an expanded version released in July 2022 containing many more labeled images and many more unlabeled images.
Below, we summarize the two distinct versions of the dataset:
We stress that in all versions, the unlabeled set is uncurated. Neither view nor diagnostic labels are available for these studies and the label distribution of these studies is not known in advance. These unlabeled sets represent what is directly available in the electronic medical record immediately after imaging a patient.
Thus, using our dataset to test SSL methods makes it a much more authentic test of the use of unlabeled examples to improve classification than alternative datasets like SVHN or CIFAR-10, where the "unlabeled" set is created by "forgetting" known labels.
Classification Tasks
Both TMED1 and TMED2 support the same two clinically-meaningful tasks: view classification and severity diagnosis classification.
Task 1: Classify the view of an image
In echocardiography, many canonical view types are possible, each displaying distinct aspects of the heart's complex anatomy.
As part of routine clinical care, when images are taken the sonographer is intentionally capturing a specific view, but the annotation of the view type is not applied to the image or recorded in the electronic record. Thus, from raw data alone (remember, each study contains 100s of images) it is difficult to focus on a specific anatomical view of interest.
For our goal of supporting diagnosis of aortic stenosis, two kinds of views are particulary relevant: parasternal long axis (PLAX) and parasternal short axis (PSAX). Both PLAX and PSAX views are used in the routine clinical assessment of aortic valve disease, because the aortic valve’s structure and function is visible.
TMED-1 makes 3 possible view-type labels available: PLAX, PSAX, or other (meaning something else other than PLAX or PSAX).
TMED-2 has been expanded to support 2 extra view-type classes: A2C and A4C. Adding labels for A2C and A4C makes some synergy with other open-access datasets (such as Stanford's EchoNet Dynamic) possible.
| TMED-1 | TMED-2 | |
|---|---|---|
| suggested classifier output | PLAX, PSAX, or other | PLAX, PSAX, A2C, A4C, or Other |
| available view labels in dataset | PLAX, PSAX, other | PLAX, PSAX, A2C, A4C, A2C-or-A4C-or-Other |
| which images have view labels? | all images in each labeled study | some images in each labeled study |
For TMED-2, annotators were instructed to label ~5 examples of each view type (PLAX, PSAX, A2C, A4C) per study so that we could get examples of relevant views from more studies.
We repurposed some images labeled "Other" within TMED-1 for TMED-2, which is why some images are labeled "A2C-or-A4C-or-Other" in TMED-2.
Example Images
We show several high-resolution examples of each of the possible view types in TMED-1 below.
Remember that the released images are 112x112 pixels (TMED-2) or 64x64 pixels (TMED-1).
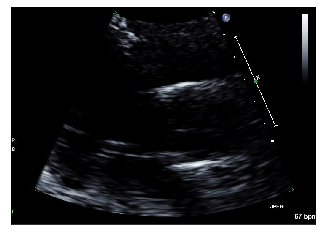
PLAX example 1/2
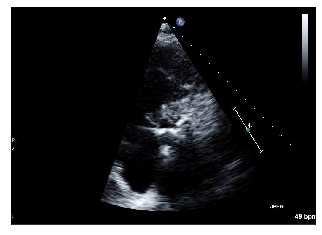
PSAX example 1/2
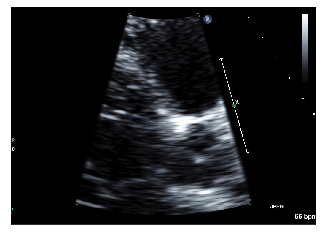
Other example 1/2
PLAX example 2/2
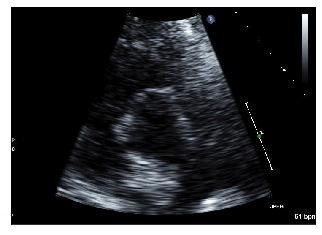
PSAX example 2/2
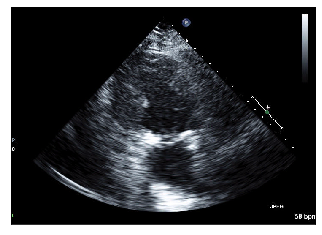
Other example 2/2
Task 2: Classify the diagnostic severity level of a patient
Our ultimate goal is automated preliminary screening of aortic stenosis (AS), which would improve early detection of this life-threatening disease.
Toward this goal, our diagnosis task requires aggregating predictions across many images of the same patient's heart (using ~100 images) to make a coherent prediction for that individual.
This tasks mimics how cardiologists make real AS diagnoses in practice: they have access to ~100 images captured by the sonographer, each of varying signal quality as well as representing different view types. The cardiologist needs to identify which images are relevant (show relevant anatomical views with appropriate quality) and then look for key signs of disease in these relevant images to determine the appropriate severity diagnosis (none, mild, moderate, or severe).
TMED-2 improves over TMED-1 by:
- making available finer-grained 5-level severity labels for each study than TMED-1
- using a more clinically-relevant grouping into 3 levels
| TMED-1 | TMED-2 | |
|---|---|---|
| suggested classifier output |
|
|
| available labels in dataset |
|
|
Acknowledgement of Funding
We gratefully acknowledge financial support from the Pilot Studies Program at the Tufts Clinical and Translational Science Institute (Tufts CTSI NIH CTSA UL1TR002544).